Decoding Human Biology & Creating Future Medicine
Welcome to the Shimizu Lab (Dept. AI Systems Medicine, Institute of Science Tokyo)

Please feel free to contact us.
Contact info: h_shimizu.dsc@@tmd.ac.jp (please remove one @)
If you are seeking a position as a graduate student, please review the information here, and send us an email complying the instructions on this page. We do not accept unscheduled visits.
For our Japanese website with more details, please visit this page.
Hideyuki SHIMIZU, MD, PhD
Physician-Scientist, Information Scientist, and Professor at Institute of Science Tokyo
He received his M.D. from Tohoku University and his Ph.D. in Medical Science from Kyushu University, JAPAN. He has held research positions at Harvard Medical School and MIT, US. His work focuses on creating the future of medicine by integrating life sciences with information technology. He currently leads over 50 collaborative projects in areas such as cancer, bioinformatics, medical AI, and synthetic biology. He holds several key appointments, including Professor at the Institute of Science Tokyo (formerly Tokyo Medical and Dental University), Program Director for the national AI talent development program (MEXT/JST BOOST), and researcher for JST's PRESTO and FOREST programs. An accomplished author and educator, he is also actively involved in media and science outreach.
We are an interdisciplinary laboratory at the cutting edge of medicine, life science, and information science. Our mission is to understand and conquer disease by deciphering complex biological data using Japan’s most powerful supercomputers, advanced AI, and data science.
Our work uniquely combines computational research with hands-on experimental validation in our own wet lab, fueled by data from leading hospitals and industry partners. We focus on elucidating disease mechanisms, developing methods for ultra-early diagnosis, and creating personalized therapies. Through AI-driven drug discovery and molecular simulations, we are designing the next generation of therapeutics and preventive medicine.
We are a highly productive, top-tier lab known for publishing in high-impact journals and fostering exceptional talent. If you are a student or postdoc passionate about shaping the future of medicine, we invite you to visit us. Let’s build the future together!

Our Team
For more information about our team, please see personal pages listed below (please note, the content is in Japanese).
- Hideyuki SHIMIZU, MD, PhD
- Satoshi OHNO, PhD
- Hidekazu HISHINUMA, PhD
- Tomoya SAKUMA, PhD candidate
- Takuya SUZUOKA, PhD candidate
- Yuki OTANI, PhD candidate
- Takumi ITO, PhD candidate
- Kazuki NAKANISHI, MD, PhD candidate
- Takashi FUJIWARA, PhD candidate
- Takahide SUZUKI, PhD candidate
- Kohei OTA, MS candidate
- Ryo OGAWA, MS candidate
- Yuto HAKATA, MS candidate
- Saki MASUDA, MS candidate
- Akinori KAWAHARADA, MD
- Naoya MINAMI, DVM candidate
- Ikuto NAKA, MD candidate
- Kazuki MATSUDA, MD candidate
- Mamoru ITOI, MD candidate
- Tae UGAKI (technical staff)
- Kazuko TANAKA (administrative staff)
We are actively seeking talented and motivated individuals to join our team.
For Prospective Graduate Students: If you are considering graduate studies with us, please review the information on this page and send the required contents to Prof. Shimizu via email.
For Postdoctoral Fellows: We welcome applications from postdoctoral researchers at any time. We especially encourage inquiries from candidates who have secured or are applying for fellowships, such as the JSPS Postdoctoral Fellowship (PD). To express your interest, please email your CV and a brief research proposal to Prof. Shimizu.
Research
Our mission is simple: to solve what is unsolvable in medicine today. We are a hybrid lab, centered on “dry” data analysis but grounded in “wet” experimental proof. We don’t just analyze data; we create new methods to prevent, diagnose, and treat disease.
Our method is a powerful three-part cycle: Measure, Model, Validate. We collaborate across disciplines—from robotics to synthetic biology—because innovation happens at the intersections.
We embrace change. We believe that flexibility in the short term makes our long-term vision stronger and our research broader. Our ultimate goal is to create the future of medicine, and to do that, we must be a team that is constantly evolving, fearless in adopting new technologies and ideas. This is challenging, but it’s also where the breakthroughs are.
We are defined by an insatiable curiosity and a relentless drive to tackle the toughest problems in medicine head-on.
A dream you dream alone is only a dream. A dream you dream together is reality.
by John Lennon
Project 1: Bioinformatics for Healthcare
Leveraging my professional expertise as a physician, our research focuses on developing analytical methods and tools to address the unique challenges presented by health and disease data. A central theme of my work is patient stratification.
Through applied research in the field of bioinformatics, we have worked to realize the promise of personalized medicine. As one example, we have developed methods to stratify cancer patient prognosis more precisely than the conventional TNM staging system.
Our research extends beyond oncology. We are also building methods to detect non-cardiac diseases solely from electrocardiogram (ECG) data and analyzing data from a diverse range of other conditions, including rheumatism, tuberculosis, and viral infections.
Precision Medicine Comes of Age
References
- Hozumi H et al., Bayesian network enables interpretable and state-of-the-art prediction of immunotherapy responses in cancer patients. PNAS Nexus 2023
- Shimizu H, Nakayama KI. A universal molecular prognostic score for gastrointestinal tumors. NPJ Genom. Med. 2021
- Aso H et al. Multiomics investigation revealing comprehensive characteristics of HIV-1-infected cells in vivo. Cell Rep. 2020
- Shimizu H, Nakayama KI. A 23 gene-based molecular prognostic score precisely predicts overall survival of breast cancer patients. EBioMedicine 2019
- Shimizu H, Nakayama KI. Artificial intelligence in oncology. Cancer Sci. 2020
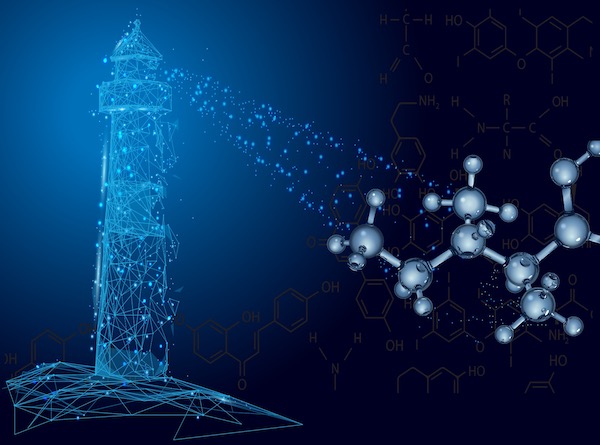
Project 2: AI DRUG Discovery
We have developed LIGHTHOUSE, an AI-powered drug discovery system designed to “illuminate” target compounds from a vast “sea” of chemical structures, often with very few initial clues. This work has been featured prominently in national newspapers and on television.
Currently, we are further expanding LIGHTHOUSE into a foundational drug discovery platform that incorporates the principles of physical chemistry. We are also tackling the complex challenge of designing middle molecule therapeutics, which offer higher specificity and affinity than conventional small molecule drugs.
In parallel, we are working to establish methods for the precise simulation of a drug’s behavior within the body, the prediction of its potential side effects, and—crucially—the development of interpretable models to understand the underlying molecular interactions.
QUEST for SMARTER THERAPEUTICS
References and media
- Sakuma T et al., High-Resolution Mapping of the Human E3-Substrate Interactome using Ubicon Uncovers Network Architecture and Cancer Vulnerabilities. bioRxiv 2025
- Otani H et al. , BaCNet: Deep Learning Accelerates Novel Antibiotic Discovery Against Resistant Pathogens. bioRxiv 2025
- Shimizu H et al. , LIGHTHOUSE illuminates therapeutics for a variety of diseases including COVID-19. iScience 2022
- NHK news
- RKB news
- Asahi newspaper
- Yomiuri newspaper
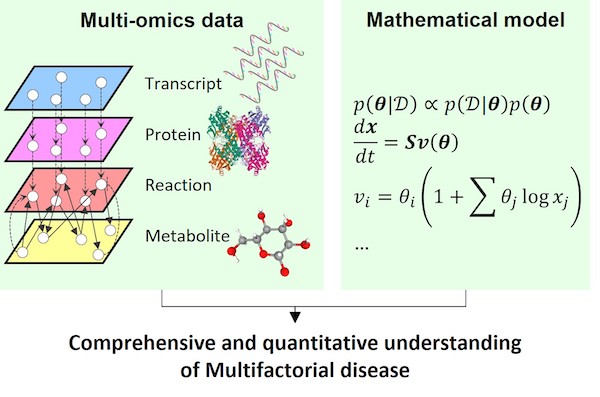
Project 3: Systems and mathematical Medicine
We approach disease as a breakdown of complex biological systems. By applying mathematical and informational analysis to large-scale measurement data, we delve deeply into understanding the pathophysiology of these conditions. Our methodology is rooted in a systems biology approach, utilizing differential equations and other advanced techniques from applied mathematics. As one example, we search for vulnerabilities in cancer—its so-called “Achilles’ heel”—to propose more effective combination therapies. In another project, we are working to decipher the precise and robust mechanisms of the immune system, incorporating perspectives from control engineering. Furthermore, we are currently constructing whole-cell simulation systems for specific diseases and cell types. We envision a future where our creations, such as the iHepatocyte, become indispensable tools for disease research and single-cell data analysis, much in the same way the iPhone revolutionized the telephone.
artificial Cell for biomedicine
References
- Ito T et al., MetDeeCINE: Deciphering Metabolic Regulation through Deep Learning and Multi-Omics. bioRxiv 2025
- Hishinuma H et al., Data-driven discovery and parameter estimation of mathematical models in biological pattern formation. PLoS Comput. Biol. 2025
- Kodama, Oshikawa, Shimizu H et al., A shift in glutamine nitrogen metabolism contributes to malignant progression of cancer. Nature Commun. 2020
PROJECT 4: Biologically Inspired computation
The field of life science has always progressed in rapid leaps with the advent of transformative technologies, from gene cloning, sequencing, and PCR to RNAi, iPS cells, genome editing, optogenetics, next-generation sequencing, and single-cell analysis. We believe that continued investment in biotechnology is essential for driving further advances in life science and, ultimately, translating them into medicine. After establishing my own lab at Tokyo Medical and Dental University, I initiated a new research program focused on DNA computing as a next-generation biotechnology. As one example, we are developing pattern recognition systems and convolutional neural networks that operate inside living cells, not on a conventional computer. By extending this technology, we aim to establish revolutionary cell therapies. While it may sound like science fiction today, we envision a future where engineered cells patrol the body to identify diseases. In the long run, our goal is to install even more complex circuits into these cells, creating stable cell lines that can serve as factories to mass-produce drugs when and where they are needed.
Any sufficiently advanced technology is indistinguishable from magic

Project Green
We call this initiative Project GREEN, for a simple reason: DNA computers require significantly less energy than conventional computers. In an era where the importance of zero-carbon initiatives and Sustainable Development Goals (SDGs) is paramount, we aim to establish an earth-friendly computing paradigm for the future by harnessing the power of biology. Of course, our ultimate goal lies beyond the "Green" aspect—it is the medical innovation this technology will unlock. A journey of a thousand miles begins with a single step, and we are diligently working to turn this grand, science-fiction-like vision into reality.PROJECT 5: Biologically Inspired computation
We are exploring the design principles of the natural world by data-mining the properties of microorganisms.
Leveraging these principles, we have recently embarked on an ambitious new theme: to create what we call “designer bacteria.” These are microorganisms engineered to efficiently produce useful compounds, with the ultimate goal of contributing to the treatment of infectious diseases and a wide range of other illnesses.
We believe in the immense potential of integrating information science and synthetic biology.
Biology by design
References
- Ito T et al. A Functional Atlas of the Tardigrade Resistome Reveals a Diverse Molecular Toolkit for Extremotolerance. bioRxiv 2025
- Otani Y et al. AMP-Atlas: Comprehensive Atlas of Antimicrobial Peptides to Combat Multidrug-resistant Bacteria. bioRxiv 2025
PROJECT 6: Quantum Computing for Biomedicine
Some problems in life science and medicine are so complex that they remain unsolvable even with today’s most powerful supercomputers. We believe the key to unlocking these challenges lies in quantum information science.
Our work focuses on developing the quantum algorithms and quantum machine learning (QML) models that will power future biomedical breakthroughs. While the hardware is still evolving, we are creating the software and theoretical frameworks that will harness its potential. For example, quantum chemistry holds the promise to revolutionize drug discovery, and we are building the tools to make that a reality.
Our ultimate goal is to pioneer innovations for tasks once thought impossible, establishing quantum information science as the next killer technology for medicine.
Quantum informatics coupled with AI accerates biomedical innovation

Are You a Challenger Ready to Define the Future of Quantum x AI?
The quantum computing era is dawning. While the hardware race requires immense resources, the future will be defined by software—by the brilliant minds who create the algorithms. This is where the true opportunities lie. It’s a field where your ideas can change everything. We have recently launched our own initiative in this space. This means you won't be joining a project with a long history, but one where you can make your mark from day one. We are looking for pioneers who will combine our lab's unique expertise in medicine and biology with the power of quantum information science. Biotech, Quantum, AI, Next-Generation Medicine: these are the fields our government is betting on for the future. In our lab, you won't have to choose. You will master them all. Join us and let's build the future together.Publications and Talks
【Original Research】
- Hishinuma H, Takigawa-Imamura H, Miura T. Data-driven discovery and parameter estimation of mathematical models in biological pattern formation. PLoS Comput. Biol. 2025 (IF=3.8)
- Fujiwara T, Kariya Y, Kobayashi K, Matsui S, Takada T. Utility of the Continuous Spectrum Formed by Pathological States in Characterizing Disease Properties. NPJ Syst. Biol. Appl. 2025 (IF=3.5)
- Koga D et al., submitted
【Preprints】
- Otani Y, Koga D, Wakizaka Y, Shimizu H. BaCNet: Deep Learning Accelerates Novel Antibiotic Discovery Against Resistant Pathogens. bioRxiv 2025
- Otani Y, Koga D, Wakizaka Y, Shimizu H. AMP-Atlas: Comprehensive Atlas of Antimicrobial Peptides to Combat Multidrug-resistant Bacteria. bioRxiv 2025
- Ito T, Ohno S, Wang Y, Uematsu S, Kuroda S, Shimizu H. MetDeeCINE: Deciphering Metabolic Regulation through Deep Learning and Multi-Omics. bioRxiv 2025
- Sakuma T, Otani Y, Shimizu H. High-Resolution Mapping of the Human E3-Substrate Interactome using Ubicon Uncovers Network Architecture and Cancer Vulnerabilities. bioRxiv 2025
- Ito T, Ota K, Hishinuma H, Shimizu H. A Functional Atlas of the Tardigrade Resistome Reveals a Diverse Molecular Toolkit for Extremotolerance. bioRxiv 2025
【Books (including Japanese) 】
- 植田充美 企画、増田咲希, 佐久間智也, 大谷悠喜, 清水秀幸分担著書、「AIとバイオの融合最前線」, シーエムシー出版 2024
【Invited talks (including Japanese) 】
- Hideyuki SHIMIZU, Unlocking Novel Antibiotics: Exploring the Latent Space with Artificial Intelligence. Pacifichem, 2025
- 清水秀幸. 機械学習による抗生物質設計. 第15回CSJ化学フェスタ2025, 2025, 船堀
- 清水秀幸. 抗菌活性を題材としたAI駆動型の天然物マイニングと中分子デザイン. CBI学会2025年大会, 2025, 船堀
- 清水秀幸. AI駆動型システム生物学による生命設計原理の解読と創薬科学への応用. 第75回 日本薬学会関西支部総会・大会, 2025, 京都
- 清水秀幸. Leveraging Machine Learning and Omics Data for Future Medicine. APPW2025(第130回日本解剖学会/第102回日本生理学会/第98回日本薬理学会合同大会), 2025
- 清水秀幸. 情報科学が加速する次世代の創薬. CPHI Japan 2025 (第22回 国際医薬品開発展)
- 清水秀幸. データ科学が駆動する創薬・医科学研究. FUJITA ICBSセミナー, 2025
- 清水秀幸. 創薬の観点からみたAI活用の現況とAIネイティブな未来の医療者育成への取り組み. 第25回日本歯科医学会学術大会, 2025
【Fellowships (including Japanese) 】
- 中西一貴. Science Tokyo CS (4年間)
- 伊東巧. Science Tokyo CS (4年間)
- 佐久間智也. Science Tokyo BOOST (3年間)
- 大谷悠喜. Science Tokyo SPRING (4年間)
- 伊東巧. Science Tokyo SPRING (4年間)
【International conference】
- Otani Y, Shimizu H. Development of an AI-driven next-generation drug discovery system for infectious diseases. Pacifichem, 2025
- Sakuma T, Shimizu H. Development of an AI-driven drug discovery model targeting G-protein-coupled receptors. Pacifichem, 2025
【Domestic conference (including Japanese) 】
- 川原田明徳, 清水秀幸. 3D構造を利用した深層学習による複数の薬物-標的間結合活性の同時予測. 第48回日本分子生物学会年会(MBSJ2025), 2025, 横浜
- 小川諒, 鈴岡拓也, 大谷悠喜, 清水秀幸. 量子化学的知見を用いたAI創薬:物理的妥当性に基づく化合物-タンパク質相互作用予測の汎化性向上に向けて. 第48回日本分子生物学会年会(MBSJ2025), 2025, 横浜
- 菱沼秀和, 清水秀幸. RNAの半減期を操り合成遺伝子回路を精密制御するRNA設計基盤の構築. 第48回日本分子生物学会年会(MBSJ2025), 2025, 横浜
- 中西一貴, 清水秀幸. AIによるTCR結合解析と自己免疫疾患に対する新薬の創出. 第48回日本分子生物学会年会(MBSJ2025), 2025, 横浜
- 佐久間智也, 清水秀幸. 深層学習を用いた核酸医薬開発の加速化. 第48回日本分子生物学会年会(MBSJ2025), 2025, 横浜
- 大田航平, 古賀大介, 麻生啓文, 清水秀幸. 多段階AIフレームワークによる抗がん剤多剤併用効果の予測. 第48回日本分子生物学会年会(MBSJ2025), 2025, 横浜
- 鈴木崇英, 白川久志, 井上飛鳥, 清水秀幸. ドラッグリポジショニングを目指したマルチモーダル・知識グラフの構築. 第48回日本分子生物学会年会(MBSJ2025), 2025, 横浜
- 鈴岡拓也, 麻生啓文, 大野聡, 清水秀幸. 神経疾患の細胞種特異性の解明に向けた短鎖反復配列が与える遺伝子発現へのゲノムワイドな影響の解析. 第48回日本分子生物学会年会(MBSJ2025), 2025, 横浜
- 藤原嵩士, 清水秀幸. 抗体-抗原相互作用予測モデルの開発と創薬への応用. 第48回日本分子生物学会年会(MBSJ2025), 2025, 横浜
- 大谷悠喜, 古賀大介, 宇垣多恵, 脇阪康成, 清水秀幸. 創薬AI「AMP-Atlas」による中分子抗菌ペプチドの網羅的探索と進化的最適化. 第48回日本分子生物学会年会(MBSJ2025), 2025, 横浜
- 伊東巧, 大野聡, 王一然, 植松沙織, 黒田真也, 清水秀幸. MetDeeCINE: 定量的代謝ネットワーク構築のためのマルチオミクス統合AI. 2025年日本バイオインフォマティクス学会年会, 2025, 名古屋
- 鈴岡拓也, 清水秀幸. メタラーニングを用いた少数データ環境下での ADMET 予測フレームワークの構築. 2025年日本バイオインフォマティクス学会年会, 2025, 名古屋
- 大田航平, 古賀大介, 麻生啓文, 清水秀幸. 抗がん剤の多剤併用効果予測のための多段階深層学習フレームワーク. 2025年日本バイオインフォマティクス学会年会, 2025, 名古屋
- 藤原嵩士, 清水秀幸. 言語モデルによるscFv-抗原相互作用の解読. 2025年日本バイオインフォマティクス学会年会, 2025, 名古屋
- 中西一貴, 清水秀幸. 対照学習を活用したTCR-pMHCクラスII結合予測AIモデルの開発. 2025年日本バイオインフォマティクス学会年会, 2025, 名古屋
- 鈴木崇英, 白川久志, 井上飛鳥, 清水秀幸. ドラッグリポジショニングを目指したマルチモーダル・知識グラフの構築. 2025年日本バイオインフォマティクス学会年会, 2025, 名古屋
- 佐久間智也, 清水秀幸. 機械学習によるGPCRを標的とした創薬の高速化. 日本薬学会第145年会, 2025
【Guest column (including Japanese) 】
- 清水秀幸. AI時代の生命科学と「知」の在り方. 文藝春秋 2025年6月号 巻頭随筆
【Original Research】
- Konno Y, Uriu K, Chikata T, Takada T, Kurita JI, Ueda MT, Islam S, Yang Tan BJ, Ito J, Aso H, Kumata R, Williamson C, Iwami S, Takiguchi M, Nishimura Y, Morita E, Satou Y, Nakagawa S, Koyanagi Y, Sato K. Two-step evolution of HIV-1 budding system leading to pandemic in the human population. Cell Rep., 43(2), 113697, 2024 (IF=10.0)
- Bai Y, Morita K, Kokaji T, Hatano A, Ohno S, Egami R, Pan Y, Li D, Yugi K, Uematsu S, Inoue H, Inaba Y, Suzuki Y, Matsumoto M, Izumi Y, Bamba T, Hirayama A, Soga T, Kuroda S. Trans-omic analysis reveals opposite metabolic dysregulation between feeding and fasting in liver associated with obesity. iScience, 27(3):109121, 2024 (IF=5.8)
- Sakuma T, Mukai Y, Yamaguchi A, Suganuma Y, Okamoto K, Furugen A, Narumi K, Ishikawa S, Saito Y, Kobayashi M. Monocarboxylate Transporters 1 and 2 Are Responsible for L-Lactate Uptake in Differentiated Human Neuroblastoma SH-SY5Y Cells. Biol. Pharm. Bull. 47(4):764-770, 2024 (IF=2.0)
- Kasai S , Shiomi A, Shimizu H, Aoba M, Kinugasa Y, Miura T, Uehara K, Watanabe J, Kawai K, Ajioka Y. Risk factors and development of machine learning diagnostic models for lateral lymph node metastasis in rectal cancer: a multicentre study. BJS Open, 2024 (IF=3.2)
- Koga D, Nakayama S, Higa T, Nakayama KI. Identification and characterization of an enhancer element regulating expression of Cdkn1c (p57 gene). Genes Cells, 2024 (IF=1.3)
【Patent (including Japanese)】
- 山田 哲也, 清水 秀幸, 小宮 力, 大野 聡, 古賀 大介, 兼田 稜. 特願2024-166123 血糖上昇の有無を予測するための装置
【Awards (including Japanese)】
- 大谷悠喜. TMDU大学院リトリート リトリート賞 (東京医科歯科大学)
- 大田航平. TMDU大学院リトリート 優秀ポスター賞 (東京医科歯科大学)
【Books (including Japanese)】
- 岡田随象 企画、川原田 明徳, 大谷 悠喜, 清水 秀幸分担著書、医学のあゆみ増刊号「遺伝統計学の新潮流 ─新規創薬・個別化医療への挑戦」, 医歯薬出版 2024
- 伊東 巧, 大田航平, 大谷 悠喜, 清水 秀幸分担著書、消化器病学サイエンス 2024年9月号「消化器領域の研究においてAIはどこまで活用されるのか?」, 先端医学社 2024
【Invited talks (including Japanese)】
- 清水 秀幸. Leveraging Machine Learning and Omics Data for Personalized Medicine. 第69回日本人類遺伝学会, 2024
- 清水 秀幸. 人工知能が創出する未来医療. 第45回日本炎症・再生医学会, 2024
- 清水 秀幸. 情報科学が創出する未来医療. 日本情報オリンピック春季トレーニング (日本代表選考会) 講義, 2024
- 清水秀幸. データサイエンスが拓く未来の生命科学と医療. 都立日比谷高校 SSH生物特別講演, 2024
【Fellowships (including Japanese)】
- 佐久間智也. TMDU-SPRING (東京医科歯科大学, 4年間)
【Meida (including Japanese)】
【International conference】
- Aso H, Shimizu H. Development of an antimicrobial resistance tracer for pandemic preparedness. CSHL Biological Data Science 2024
- Ito T, Ohno S, Wang Y, Kuroda S, Shimizu H. Metabolic regulatory network reconstruction using artificial intelligence and multi-omic data. Asia & Pacific Bioinformatics Joint Conference 2024
- Otani Y, Wakizaka Y, Koga D, Aso H, Shimizu H. AI-assisted discovery of antimicrobial peptides to combat multidrug-resistant bacteria. Asia & Pacific Bioinformatics Joint Conference 2024
【Domestic conference (including Japanese)】
- 笠井俊輔、塩見明生、青葉萌奈美、清水秀幸、賀川弘康、日野仁嗣、眞部祥一、山岡雄祐、前田周良、田中佑典、川合一茂、味岡洋一. 直腸癌側方リンパ節転移診断能向上を目指した機械学習モデルの開発. 第100回大腸癌研究会学術集会, 2024
- 笠井俊輔、塩見明生、青葉萌奈美、清水秀幸、賀川弘康、眞部祥一、山岡雄祐、前田周良、川合一茂、味岡洋一. 直腸癌側方リンパ節転移診断能向上を目指した機械学習モデルの開発. 第79回日本消化器外科学会総会, 2024
- 大谷悠喜、脇阪康成、古賀大介、麻生啓文、清水秀幸. AI駆動の次世代感染症創薬システムの開発. 第47回日本分子生物学会年会, 2024
- 佐久間智也、麻生啓文、古賀大介、清水秀幸. 核酸医薬開発加速化のための大規模言語モデルを活用した機械学習モデルの開発. 第47回日本分子生物学会年会, 2024
- 大野聡、清水秀幸. シングルセルフラクソミクスに向けて. 第47回日本分子生物学会年会, 2024
- 鈴岡拓也、麻生啓文、古賀大介、清水秀幸. 合理的な薬剤設計のための薬物動態を考慮した ADMET 予測AIの開発. 第47回日本分子生物学会年会, 2024
- 大田航平、古賀大介、麻生啓文、清水秀幸. AIによる抗がん剤多剤併用療法の効果予測. 第47回日本分子生物学会年会, 2024
- 増田咲希、大野聡、古賀大介、大谷悠喜、大田航平、伊東巧、清水秀幸. 代謝疾患の定量的な理解に向けた酵素パラメータ予測AIの開発. 第47回日本分子生物学会年会, 2024
- 博田悠斗、大谷悠喜、麻生啓文、古賀大介、清水秀幸. 神経疾患に対する治療薬開発のための血液脳関門透過ペプチドをデザインするAIの開発. 第47回日本分子生物学会年会, 2024
- 小川諒、大野聡、佐久間智也、伊東巧、大田航平、清水秀幸. 急性骨髄性白血病の統合ネットワーク解析. 第47回日本分子生物学会年会, 2024
- 沖田大和, 坂東希歩, 山崎美紅, 天羽宏枝, 大野聡, 清水秀幸, 鬼塚正義. 計算化学技術を活用した二重特異性抗体の凝集性の改善. 第3回日本抗体学会学術大会, 2024
【Original Research】
- Hozumi, Shimizu H. Bayesian network enables interpretable and state-of-the-art prediction of immunotherapy responses in cancer patients. PNAS Nexus, 2(5), pgad133, 2023
- Fujinuma S, Nakatsumi H, Shimizu H, Sugiyama S, Harada A, Goya T, Tanaka M, Kohjima M, Takahashi M, Izumi Y, Yagi M, Kang D, Kaneko M, Shigeta M, Bamba T, Ohkawa Y, Nakayama KI. FOXK1 promotes nonalcoholic fatty liver disease by mediating mTORC1-dependent inhibition of hepatic fatty acid oxidation. Cell Rep., 42(5), 112530, 2023 (IF=10.0)
- Oka T, Higa T, Sugahara O, Koga D, Nakayama S, Nakayama KI. Ablation of p57+ quiescent cancer stem cells suppresses recurrence after chemotherapy of intestinal tumors. Cancer Res., 2023 (IF=13.3)
- Hirooka K, Oki K, Ogawa-Ochiai K, Nakaniida Y, Onoe H, Kiuchi Y. Blood flow in the optic nerve head in patients with primary aldosteronism. PLoS One, 18(4), e0285039, 2023 (IF=3.7)
- Kodama M, Toyokawa G, Sugahara O, Sugiyama S, Haratake N, Yamada Y, Wada R, Takamori S, Shimokawa M, Takenaka T, Tagawa T, Kittaka H, Tsuruda T, Tanaka K, Komatsu Y, Nakata K, Imado Y, Yamazaki K, Okamoto I, Oda Y, Takahashi M, Izumi Y, Bamba T, Shimizu H, Yoshizumi T, Nakayama KI. Modulation of host glutamine anabolism enhances the sensitivity of small cell lung cancer to chemotherapy. Cell Rep., 42(8):112899, 2023 (IF=10.0)
- Nakaniida Y, Tokumo K, Onoe H, Okada N, Nakakura S, Asaoka R, Kiuchi Y. Comparison of Corvis ST Parameters between Primary Open-Angle Glaucoma and Primary Angle-Closure Glaucoma. J. Clin. Med., 12(15), 5108, 2023 (IF=4.9)
- Aso H, Ito J, Ozaki H, Kashima Y, Suzuki Y, Koyanagi Y, Sato K. Single-cell transcriptome analysis illuminating the characteristics of species-specific innate immune responses against viral infections. GigaScience, 12, 2023 (IF=7.6)
- Yamaguchi A, Mukai Y, Sakuma T, Suganuma Y, Furugen A, Narumi K, Kobayashi M. Molecular characteristic analysis of single-nucleotide polymorphisms in
SLC16A9/hMCT9. Life Sci., 334, 122205, 2023 (IF=6.1) - Funasaki S, Hatano A, Nakatsumi H, Koga D, Sugahara O, Yumimoto K, Baba M, Matsumoto M, Nakayama KI. A stepwise and digital pattern of RSK phosphorylation determines the outcome of thymic selection. iScience 26 (9), 107552, 2023 (IF=5.8)
- Nakaniida Y, Higashikawa F, Tokumo K, Yuasa Y, Onoe H, Okada N, Nakakura S, Asaoka R, Kiuchi Y. Cureus 15(10): e46500, 2023 (IF=1.2)
- Maehara H, Kokaji T, Hatano A, Suzuki Y, Matsumoto M, Nakayama KI, Egami R, Tsuchiya T, Ozaki H, Morita K, Shirai M, Li D, Terakawa A, Uematsu S, Hironaka K, Ohno S, Kubota H, Araki H, Miura F, Ito T, Kuroda S. DNA hypomethylation characterizes genes encoding tissue-dominant functional proteins in liver and skeletal muscle. Sci. Rep., 2023 (IF=5.0)
- Yamaguchi A, Mukai Y, Sakuma T, Furugen A, Narumi K, Kobayashi M. Atorvastatin Exerts More Selective Inhibitory Effects on hMCT2 than on hMCT1 and hMCT4. Anticancer Res., 43(7):3015-3022, 2023 (IF=2.0)
- Yamaguchi A, Mukai Y, Sakuma T, Narumi K, Furugen A, Yamada Y, Kobayashi M. Monocarboxylate transporter 4 involves in energy metabolism and drug sensitivity in hypoxia. Sci. Rep., 13(1):1501, 2023 (IF=4.6)
【Awards (including Japanese)】
- 清水秀幸. 第27回丸文研究奨励賞 (一般財団法人 丸文財団)
- 大谷悠喜. MBSJ2023サイエンスピッチ優秀発表賞 (日本分子生物学会)
【Books (including Japanese)】
- 清水 秀幸 編著. 実験医学別冊 Pythonで実践 生命科学データの機械学習. 羊土社 2023
- 大澤 毅 編、大野 聡 分担著書、実験医学増刊「マルチオミクス データ駆動時代の疾患研究」. 羊土社 2023
- 「一家に一枚 ウイルス」製作チーム (小嶋将平、渡士幸一、渡辺登喜子、堀江真行、麻生啓文、多賀佳、川久保修介、塩野谷果歩) 製作監修. 学習資料「一家に1枚 ウイルス」 文部科学省 2023
【Meida (including Japanese)】
- 清水 秀幸. 感染症TODAY (ラジオNIKKEI社の全国放送, 医学番組) 出演
【Invited talks (including Japanese)】
- 清水 秀幸. ネクストパンデミックを数理情報科学との融合知で防ぐ. TMDU感染症センター記念シンポジウム, 2023
- Hideyuki Shimizu. Toward personalized cancer treatment through AI drug discovery. 第82回日本癌学会学術総会, シンポジウム, 2023
- Hideyuki Shimizu. Toward ultra-fast drug discovery for hundreds of pathogens. 日米先端工学シンポジウム (JAFOE) シンポジウム, 2023
- 清水 秀幸. 医療とAI入門. 日本脳神経血管内治療学会関東地方会学術集会, 2023
- Hideyuki Shimizu. Future challenges of AI in oncology. Chugai Cancer Summit, 2023 (欧州の医師やがん研究者, 専門家を対象にした招待講演とパネルディスカッション)
- 清水 秀幸. 臨床応用を目指したデータ科学・AI創薬研究. 東北大学主催第1回医学AIセミナー, 2023
- 清水 秀幸. データサイエンスによる小細胞肺がん治療法の確立を目指して. 15th NUJRA special seminar, 2023
- 清水 秀幸. オミクス解析における人工知能技術の活用. 千里ライフサイエンスセミナーT5 「バイオインフォマティクスの最前線」, 2023
- 清水 秀幸. オンラインによる学生医学研究・交流のすゝめ, 第31回日本医学会総会学生企画, 2023
- 清水 秀幸. 医療におけるAIの活用と課題, LabTech Talk, 2023
- 清水 秀幸. 異分野融合で創り出す未来医療~新大学で目指すConvergence Scienceとは何だろう?~ 第72回お茶の水祭, 2023
【International conference】
- Nakaniida Y et al. Characteristics of Corvis ST parameters in subjects with low ocular blood flow. The 38th Asia-Pacific Academy of Ophthalmology (APAO) Congress. Kuala Lumpur, Malaysia, 2023
- Ohno S et al., OMELET: Multi-omics-based metabolic flux inference for understanding obesity-associated dysregulatory mechanisms in liver glucose metabolism. Metabolic Engineering 15. Singapore, 2023
【Domestic conference (including Japanese)】
- 麻生 啓文, 伊東 潤平, 尾崎 遼, 鹿島 幸恵, 鈴木 穣, 小柳 義夫, 佐藤 佳. ウイルス感染に対する動物種特異的自然免疫応答. 第17回ウイルス学キャンプ in 湯河原, 2023
- 中新井田 悠太, 徳毛 花菜,尾上 弘光,岡田 尚樹, 中倉 俊祐,朝岡 亮, 清水 秀幸, 木内良明. 原発開放隅角緑内障と原発閉塞隅角緑内障の Corvis ST パラメータの比較. 第77回日本臨床眼科学会, 2023
- 中新井田 悠太, 清水 秀幸, 大野 聡, 徳毛 花菜, 尾上 弘光, 岡田 尚樹, 中倉 俊祐, 朝岡 亮, 木内 良明. 機械学習を用いた緑内障・白内障同時手術後の視力予測. 第4回日本眼科AI学会総会, 2023
- 麻生 啓文, 大野 聡, 清水 秀幸. プラスミドDNAの宿主細菌を予測するツールの開発. 第46回日本分子生物学会年会, 2023
- 伊東 巧, 大野 聡, Yiran Wang, 黒田 真也, 清水 秀幸. AIとマルチオミクスデータを用いたデータドリブンな代謝ネットワークの再構築. 第46回日本分子生物学会年会, 2023
- 大谷 悠喜, 脇阪 康成, 麻生 啓文, 古賀 大介, 清水 秀幸. 微生物学を学んだAIによる次世代の感染症創薬に向けて. 第46回日本分子生物学会年会, 2023
- 古賀 大介, 麻生 啓文, 大野 聡, 清水 秀幸. 機械学習を用いたゲノミクスによる2型糖尿病の病態解析. 第46回日本分子生物学会年会, 2023
- 大田 航平, 古賀 大介, 麻生 啓文, 清水 秀幸. 抗がん剤の多剤併用療法の効果予測モデルの開発. 第46回日本分子生物学会年会, 2023
- 佐久間 智也, 向井悠斗, 山口敦史, 石川修平, 古堅彩子, 鳴海克哉, 小林正紀. 分化誘導されたヒト神経芽細胞腫SH-SY5Y細胞における乳酸輸送担体の同定. 日本薬学会第143年会, 2023
【Original Research】
- Mise S, Matsumoto A, Shimada K, Hosaka T, Takahashi M, Ichihara K, Shimizu H, Shiraishi C, Saito D, Suyama M, Yasuda T, Ide T, Izumi Y, Bamba T, Kimura-Someya T, Shirouzu T, Miyata H, Ikawa M, Nakayama KI. Kastor and Polluks polypeptides encoded by a single gene locus cooperatively regulate VDAC and spermatogenesis. Nature Commun., 13(1), 1071, 2022 (IF=17.7)
- Uematsu S, †Ohno S, Tanaka KY, Hatano A, Kokaji T, Ito Y, Kubota H, Hironaka K, Suzuki Y, Matsumoto M, Nakayama KI, Hirayama A, Soga T, †Kuroda S. Multi-omics-based label-free metabolic flux inference reveals obesity-associated dysregulatory mechanisms in liver glucose metabolism. iScience, 25(2), 103787. (2022) (IF=6.1) (†corresponding authors)
- Habara M, Sato Y, Goshima T, Sakurai M, Imai H, Shimizu H, Katayama Y, Hanaki S, Masaki T, Morimoto M, Nishikawa S, Toyama T, Shimada M. FKBP52 and FKBP51 Differentially Regulate the Stability of Estrogen Receptor in Breast Cancer. Proc Natl Acad Sci USA, 19(15), e2110256119, 2022 (IF=11.2)
- Terakawa A, Hu Y, Kokaji T, Yugi K, Morita K, Ohno S, Pan Y, Bai Y, Parkhitko AA, Ni X, Asara JM, Bulyk ML, Perrimon N, Kuroda S. Trans-omics analysis of insulin action reveals a cell growth subnetwork which co-regulates anabolic processes. iScience, 25(5), 104231, 2022 (IF=6.1)
- †Shimizu H, Kodama M, Matsumoto M, Orba Y, Sasaki Y, Sato A, Sawa H, †Nakayama KI. LIGHTHOUSE illuminates therapeutics for a variety of diseases including COVID-19. iScience, 25(11), 105314, 2022 (IF=6.1) (†corresponding authors)【解説記事はこちら】
- Kokaji T, Eto M, Hatano A, Yugi K, Morita K, Ohno S, Fujii M, Hironaka K, Ito Y, Egami R, Uematsu S, Terakawa A, Pan Y, Maehara H, Li D, Bai Y, Tsuchiya T, Ozaki H, Inoue H, Kubota H, Suzuki Y, Hirayama A, Soga T, Kuroda S. In vivo transomic analyses of glucose-responsive metabolism in skeletal muscle reveal core differences between the healthy and obese states. Sci. Rep. 12(1), 1–19, 2022 (IF=5.0)
- Yamato M, Matsuyama S, Murakami Y, Aida J, Lu Y, Sugawara Y, Tsuji I. Association between the number of remaining teeth and disability-free life expectancy, and the impact of oral self-care in older Japanese adults: a prospective cohort study. BMC Geriatr. 22(1), 820, 2022 (IF=3.7)
- Huicheng Q, Kikuchi M, Yoshino Y, Fang Z, Ohashi K, Gotoh T, Ideta R, Ui A, Endo S, Otsuka K, Shindo N, Gonda K, Ishioka C, Chiba N. BRCA1 transports the DNA damage signal for CDDP-induced centrosome amplification through the centrosomal Aurora A. 13(12), 4230-4243, Cancer Sci., 2022 (IF=6.5)
- Higa T, Okita Y, Matsumoto A, Nakayama S, Oka T, Sugahara O, Koga D, Takeishi S, Nakatsumi H, Hosen N, Robine S, Taketo MM, Sato T, Nakayama KI. Spatiotemporal reprogramming of differentiated cells underlies regeneration and neoplasia in the intestinal epithelium. Nature Commun., 13(1), 1500, 2022 (IF=17.7)
- Mukai Y, Yamaguchi A, Sakuma T, Nadai T, Furugen A, Narumi K, Kobayashi M. Involvement of SLC16A1/MCT1 and SLC16A3/MCT4 in l-lactate transport in the hepatocellular carcinoma cell line. Biopharm. Drug Dispos. 43(5):183-191, 2022 (IF=2.1)
【Review】
- †Ohno S, Uematsu S, Kuroda S. Quantitative metabolic fluxes regulated by trans-omic networks. Biochem. J. 479(6), 787–804, 2022 (IF=3.8) (†corresponding author)
【Invited talks (including Japanese)】
- 清水 秀幸. Machine learning-guided drug discovery: Beyond protein structures, 第45回日本分子生物学会年会ワークショップ, 2022
- 大野 聡, 黒田真也. 少数の酵素に対するインスリン依存性リン酸化とアロステリック制御による脂肪細胞の糖代謝制御. 第45回日本分子生物学会年会ワークショップ, 2022
- Aso H, Ito J, Ozaki H, Kashima Y, Suzuki Y & Sato K. ウイルス感染に対する自然免疫応答の動物種間比較解析. 第69回日本ウイルス学会学術集会. 2022
- 清水 秀幸. 抗原提示の法則をデータサイエンスで紐解く, 東京医科歯科大学重点領域研究全体会議, 2022
- 清水 秀幸. 医・歯・保健領域横断的なデータサイエンス交流の始動, DSCシンポジウム, 2022
- 清水 秀幸. データサイエンスによる次世代型医療への挑戦, 東京大学理学部セミナー, 2022
【Meida (including Japanese)】
【Press release (including Japanese)】
- 2種類の小さなタンパク質が精子ミトコンドリアの形態を制御する -男性不妊のメカニズムの一端を解明-
- 乳がん治療法の開発に新たな光~プロリン異性化酵素による乳がんの悪性化機構を発見~
- 数理モデルを用いた肥満における代謝変化と制御の定量的理解
【International conference】
- Nakaniida Y, Higashikawa F, Asaoka R, Nakakura S, Kiuchi Y. Effect of Axial length on Subjective Refraction and UCVA. The 14th International Orthoptic Association Congress 2022, Virtual Meeting, 2022
- Aso H, Ito J, Ozaki H, Kashima Y, Suzuki Y & Sato K. Comparative analysis of innate immune response to viral infection among animal species. EMBO Workshop Pathogen immunity and signaling. Saint-Malo, France. 2022
【Domestic conference (including Japanese)】
- 大野 聡. 糖投与後のマウスにおける臓器連関の代謝フラックス解析. CREST多細胞 領域会議, 2022
- 大野 聡. マルチオミクスデータから代謝フラックスとその制御を解析する手法OMELETの開発, UBIセミナー, 2022
- Ohno S. Metabolic flux analysis for understanding metabolic regulation in mammalian cells and organs. CREST多細胞 rising star webinar, 2022
- 大和真弥、松山紗奈江、村上義孝、相田潤、陸兪凱、菅原由美、辻一郎. 現在歯数および口腔ケアと健康寿命との関連:大崎コホート2006研究. 第32回日本疫学会学術総会, 2022
- 中新井田 悠太, 東川史子,湯浅勇生, 徳毛花菜,尾上弘光,岡田尚樹, 中倉俊祐,朝岡亮, 木内良明. 健常者における LSFG と Corvis ST パラメータの関連性について. 第126 回日本眼科学会総会, 2022
- 中新井田 悠太, 東川史子,湯浅勇生, 徳毛花菜,尾上弘光,岡田尚樹, 中倉俊祐, 朝岡亮, 木内良明. 屈折異常が眼血流に与える影響. 第4回日本近視学会総会, 2022
- 中新井田 悠太, 東川史子,湯浅勇生, 徳毛花菜,尾上弘光,岡田尚樹, 中倉俊祐,朝岡亮, 木内良明. 第76 回日本臨床眼科学会, 2022
- Koga D, Higa T, Nakayama S, Oka T, Sugahara S, Nakayama KI. Searching for the p57 gene regulatory regions in mouse ESCs. 第45回日本分子生物学会年会, 2022
【Original Research】
- Shimizu H, Nakayama KI. A universal molecular prognostic score for gastrointestinal tumors. NPJ Genom. Med., 6(1), 6, 2021 (IF=8.6)
- Egami R, Kokaji T, Hatano A, Yugi K, Eto M, Morita K, Ohno S, Fujii M, Hironaka K, Uematsu S, Terakawa A, Bai Y, Pan Y, Tsuchiya T, Ozaki H, Inoue H, Uda S, Kubota H, Suzuki Y, Matsumoto M, Nakayama KI, Hirayama A, Soga T, Kuroda S. Trans-omic analysis reveals obesity-associated dysregulation of inter-organ metabolic cycles between the liver and skeletal muscle. iScience, 24(3), 102217. (2021) (IF=6.1)
- Ichihara K, Matsumoto A, Nishida H, Kito Y, Shimizu H, Shichino Y, Iwasaki S, Imami K, Ishihama Y, Nakayama KI. Combinatorial analysis of translation dynamics reveals eIF2 dependence of translation initiation at near-cognate codons. Nucleic Acids Res., 49(13), 7298-7317, 2021 (IF=16.9)【解説記事はこちら】
- Kosugi Y, Uriu K, Suzuki N, Yamamoto K, Nagaoka S, Kimura I, Konno Y, Aso H, Willett BJ, Kobayashi T, Koyanagi Y, Ueda MT, Ito J, Sato K. A comprehensive investigation on the interplay between feline APOBEC3Z3 proteins and feline immunodeficiency virus Vif proteins. J. Virol., 95(13):e0017821, 2021 (IF=6.5)
【Invited talks (including Japanese)】
- 大野聡. 脂肪細胞におけるインスリン依存性糖代謝の速度論的トランスオミクス解析. 質量インフォマティクス研究会2021年ワークショップ, 2021
- 大野聡, 黒田真也. 脂肪細胞におけるインスリン依存性糖代謝の速度論的トランスオミクス解析. 日本プロテオーム学会2021年大会, 2021
【Patent (including Japanese)】
- 中山 敬一, 清水 秀幸. 特願2021-091667 相互作用推定方法、相互作用推定装置および相互作用推定プログラム
【Meida (including Japanese)】
- KBC(九州朝日放送) : 九大 人工知能で治療薬
- KBC(九州朝日放送) : 治療薬を選別するAⅠを開発 新型コロナの治療薬も?
- 日テレ: 新薬の開発期間短縮に期待 九州大学が人工知能を開発
- NHKニュース:AIで治療薬候補を高速予測“コロナ感染抑制物質も”九州大など
- 朝日新聞: 薬の候補予測 AIで速さ数千倍
- 読売新聞: 新薬候補AIが探す
- 産経新聞: AIで新薬開発加速
- 毎日新聞: 新薬開発、AIで高速化
- 西日本新聞: “新薬” 発見速度 AIで1000倍↑
- 文藝春秋オピニオン・2022年の論点100 : AIでコロナ治療薬を迅速に見つける”灯台”
- 日経デジタルヘルス : 治療薬の探索を数千倍速く、九大などの高速創薬AIがスキップした「障壁」
- マイナビニュース: AIを用いてデルタ株を含む新型コロナ治療薬候補の発見に九大が成功
【Press release (including Japanese)】
【International conference】
- Way J, Dobbertin T, Heid D, Chan KR, Vernet A, Sanchez-Ventura M, Graveline A, Burrill D, Lee J, Shimizu H, Niopek D, Silver P. Harnessing the natural trypsin antagonist SPINK-1 for targeted treatment of acute pancreatitis. 52nd Annual Meeting of the American Pancreatic Association, USA, 2021
- Nakaniida Y, Yuasa Y, Asaoka R, Nakakura S, Kiuchi Y. Comparison of Corvis ST Tonometry Parameters between Primary Open-Angle Glaucoma and Primary Angle- Closure Glaucoma. 9th World Glaucoma Congress, Virtual Meeting, 2021
- Nakaniida Y, Higashikawa F, Asaoka R, Nakakura S, Kiuchi Y. Effect of ocular dominance on fundus blood flow. The 14th Joint Meeting of Japan-China-Korea Ophthalmologists, Virtual Meeting, 2021
【Domestic conference (including Japanese)】
- Koga D, Higa T, Nakayama S, Oka T, Sugahara S, Nakayama KI. The role of transcription factor Etv4 in intestinal regeneration. 第44回日本分子生物学会年会, 2021
- 植松沙織, 大野 聡, 黒田真也. Omics-based label-free metabolic flux inference reveals dysregulation of glucose metabolism in liver associated with obesity. 2021年日本バイオインフォマティクス学会年会, 2021 【Highlight track】
【Original Research】
- Onoyama I, Nakayama S, Shimizu H, Nakayama KI. Loss of Fbxw7 impairs development of and induces heterogeneous tumor formation in the mouse mammary gland. Cancer Res., 80(24), 5515-5530, 2020 (IF=13.3) 【解説記事はこちら】
- Kokaji T, Hatano A, Ito Y, Yugi K, Eto M, Morita K, Ohno S, Fujii M, Hironaka K, Egami R, Terakawa A, Tsuchiya T, Ozaki H, Inoue H, Uda S, Kubota H, Suzuki Y, Ikeda K, Arita M, Matsumoto M, Nakayama K.I, Hirayama A, Soga T, Kuroda S. Transomics analysis reveals allosteric and gene regulation axes for altered hepatic glucose-responsive metabolism in obesity. Science Signaling, 13(660), eaaz1236. (2020) (IF=8.2)
- Yamauchi Y, Nita A, Nishiyama M, Muto Y, Shimizu H, Nakatsumi H, Nakayama KI. Skp2 contributes to cell cycle progression in trophoblast stem cells and to placental development. Genes Cells, 25(6), 427-438, 2020 (IF=2.30)
- Ohno S, Quek L-E, Krycer JR, Yugi K, Hirayama A, Ikeda S, Shoji F, Suzuki K, Soga T, James DE, Kuroda S. Kinetic trans-omic analysis reveals key regulatory mechanisms for insulin-regulated glucose metabolism in adipocytes. iScience, 23(9), 101479. (2020) (IF=6.1)
- Oshikawa K, Matsumoto M, Kodama M, Shimizu H, Nakayama KI. A fail-safe system to prevent oncogenesis by senescence is targeted by SV40 small T antigen. Oncogene, 39(10), 2170-2186, 2020 (IF=9.87)
- Quek L-E, Krycer JR, Ohno S, Yugi K, Fazakerley DJ, Scalzo R, Elkington SD, Dai Z, Hirayama A, Ikeda S, Shoji F, Suzuki K, Locasale JW, Soga T, James DE, Kuroda S. Dynamic 13C Flux Analysis Captures the Reorganization of Adipocyte Glucose Metabolism in Response to Insulin. iScience, 23(2). (2020) (IF=6.1)
- 【Citation Top1% paper】Kodama M, Oshikawa K, Shimizu H, Yoshioka S, Takahashi M, Izumi Y, Bamba T, Tateishi C, Tomonaga T, Matsumoto M, Nakayama KI. A shift in glutamine nitrogen metabolism contributes to malignant progression of cancer. Nature Commun., 11(1), 1320, 2020 (IF=14.9)【解説記事はこちら】
- Hwang YS, Suzuki S, Saita Y, Ito J, Sakata Y, Aso H, Sato K, Hermann BP, Sasaki K. Reconstitution of prospermatogonial specification in vitro from human induced pluripotent stem cells. Nature Commun.,11(1), 5656, 2020 (IF=14.9)
- Nakano Y, Keisuke Y, Ueda MT, Soper A, Konno Y, Kimura I, Uriu K, Kumata R, Aso H, Misawa N, Nagaoka S, Shimizu S, Mitsumune K, Kosugi Y, Juarez-Fernandez G, Ito J, Nakagawa S, Ikeda T, Koyanagi Y, Harris RS, Sato K. A role for gorilla APOBEC3G in shaping lentivirus evolution including transmission to humans. PLoS Pathog., 16(9), e1008812, 2020 (IF=6.8)
- Aso H, Nagaoka S, Kawakami, Ito J, Islam S, Tan BJY, Nakaoka S, Ashizaki K, Shirogushi K, Suzuki Y, Satou Y, Koyanagi Y, Sato K. Multiomics investigation revealing comprehensive characteristics of HIV-1-infected cells in vivo. Cell Rep., 32 (2), 107887, 2020 (IF=9.4)
【Review】
- 【Citation Top1% paper】Shimizu H, Nakayama KI. Artificial intelligence in oncology. Cancer Sci., 111(5), 1452-1460, 2020 (IF=6.72)
【Invited talks (including Japanese)】
- 大野 聡, 黒田真也. インスリン刺激下の脂肪細胞における速度論的トランスオミクス解析. 第51回化学工学会秋季大会, 2020
【Press release (including Japanese)】
- 難治性乳がんの原因となる腫瘍内多様性発生のメカニズムを解明 -乳がんの新たな治療戦略の開発に期待-
- がんの悪性化における窒素代謝の変化を解明 -難治性がんの新たな治療標的PPATを発見-
- インスリン作用による脂肪細胞の糖代謝制御の全貌を解明
【Domestic conference (including Japanese)】
- Sato K, Aso H, Nagaoka S, Ito J. シングルセルトランスクリプトーム解析による生体内におけるエイズウイルス感染細胞の異質性の描出. 第71回日本電気泳動学会総会, 2020
【Original Research】
- 【Citation Top10% paper】Shimizu H, Nakayama KI. A 23 gene-based molecular prognostic score precisely predicts overall survival of breast cancer patients. EBioMedicine, 46, 150-159, 2019 (IF=11.2)【解説記事はこちら】
- Muto Y, Moroishi T, Ichihara K, Nishiyama M, Shimizu H, Eguchi H, Moriya K, Koike K, Mimori K, Mori M, Katayama Y, Nakayama KI. Disruption of FBXL5-mediated cellular iron homeostasis promotes liver carcinogenesis. J. Exp. Med.,216(4), 950-965, 2019 (IF=14.3)【解説記事はこちら】
- Shimizu H, Takeishi S, Nakatsumi H, Nakayama KI. Prevention of cancer dormancy by Fbxw7 ablation eradicates disseminated tumor cells. JCI Insight, 4(4), e125138, 2019 (IF=9.5)【解説記事はこちら】
- Aso H, Ito J, Koyanagi Y, Sato K. Comparative Description of the Expression Profile of Interferon- stimulated Genes in Multiple Cell Lineages Targeted by HIV-1 Infection. Front. Microbiol., 10, 429, 2019 (IF=4.2)
【Review】
- Yugi K, Ohno S, Krycer JR, James DE, Kuroda S. Rate-oriented trans-omics: integration of multiple omic data on the basis of reaction kinetics. Curr. Opin. Syst. Biol. 15, 109–120, 2019 (IF=2.9)
【Books (including Japanese)】
- (分担執筆) 清水 秀幸, 中山 敬一. 「人と共生するAI革命―活用事例からみる生活・産業・社会の未来展望」エヌ・ティー・エス出版, 2019
【Invited talks (including Japanese)】
- Shimizu H, Nakayama KI. Machine learning uncovers novel aspects in healthcare and life science, 第42回日本分子生物学会シンポジウム, 2019
- 清水 秀幸. データ駆動科学によるがん個別化医療への試み, 東北大学加齢医学研究所セミナー, 2019
- Ohno S, Hirayama A, Soga T, James DE, Kuroda S. Regulation of metabolic fluxes in adipocytes with insulin. 第13回メタボロームシンポジウム, 2019
【Original Research Written in Japanese 】
- 中新井田 悠太, 半田知也, 川守田拓志, 岩田遥, 庄司信行. 片眼遮閉下と両眼開放下における中間距離の屈折値変化. 視覚の科学, 40(3), 51-54, 2019 【 Young investigator award / 第 25 回日本眼光学学会学術奨励賞受賞】
【Meida (including Japanese)】
【Press release (including Japanese)】
【International conference】
- Ohno S, Quek LE, Krycer JR, Yugi Y, Hirayama A, Soga T, James DE, Kuroda S. Metabolic flux changes over time and the regulatory mechanisms in insulin-stimulated adipocytes. ICSB2019, Japan, 2019
【Domestic conference (including Japanese)】
- 見世 慎太朗、松本 有樹修、市原 知哉、清水 秀幸、高橋 政友、和泉 自泰、馬場 健史、宮田 治彦、嶋田 圭祐、伊川 正人、中山 敬一. 精子特異的な2つの新規ポリペプチドはVDACの機能と精子ミトコンドリアの形態形成を制御する. 第42回 日本分子生物学会年会, 2019
- 小玉 学、押川 清孝、清水 秀幸、吉岡 進、高橋 政友、和泉 自泰、馬場 健史、朝長 毅、松本 雅記、中山 敬一. 核酸合成経路へのグルタミン代謝シフトはヒトがんの悪性化を可能とする. 第42回 日本分子生物学会年会, 2019
- 澤田 高志、清水 秀幸、山内 悠平、中山 敬一. 腫瘍進展を抑制し予後に関わるマイクロRNA miR-139の同定. 第42回 日本分子生物学会年会, 2019 【詳しくはこちら】
- 清水 秀幸, An AI-based Scoring System Precisely Predicts Overall Survival of Breast Cancer Patients. 第14回研究所ネットワーク国際シンポジウム, 2019
- Sato K, Nakano Y, Yamamoto K, Ueda MT, Soper A, Kumata R, Aso H, Misawa N, Konno Y, Kimura I, Nagaoka S, Juarez-Fernandez G, Ito J, Nakagawa S, Koyanagi Y. Gorilla APOBEC3G restricts SIVcpz and influences lentiviral evolution in great ape cross-species transmissions. The 18th Awaji International Forum on Infection and Immunity. 2019
- Aso H, Nagaoka S, Ito J, Misawa N, Shiroguchi K, Suzuki Y, Kawakami E, Koyanagi Y & Sato K. Characterization of the heterogeneity of HIV-1-infected cells in vivo. 第67回日本ウイルス学会学術集会. 2019
- Aso H, Ito J, Koyanagi Y, Sato K. Transcriptomic meta-analysis of interferon-stimulated genes in multiple cell lineages targeted by HIV-1 infection. The 17th International Student Seminar. 2019
- Nagaoka S, Aso H, Ito J, Misawa N, Islam S, Shiroguchi K, Kawakami E, Koyanagi Y & Sato K、生体内HIV-1感染細胞のマルチオミクス解析. SRC. 2019
- Aso H, Ito J, Sato K. HIV-1感染標的細胞におけるインターフェロン誘導遺伝子発現パターンの網羅的解析. 第16回ウイルス学キャンプ in 湯河原. 2019
- 大野 聡, Quek LE, Krycer JR, 柚木克之,平山明由,曽我朋義, James DE, 黒田真也. インスリン刺激下の脂肪細胞の代謝フラックスおよびその調節の数理解析. 第四回生活習慣病とがんの代謝栄養メカニズム研究会, 2019 【 Best poster award / 最優秀ポスター賞 受賞】
- 中新井田 悠太, 渋谷恵理, 清田優希也, 丸山美貴, 関枝里子, 三田哲大, 久保江 理, 佐々木洋. 術前混濁による予測視力からの焦点深度拡張型眼内レンズと二焦点眼内レンズ の適応判定の試み. 第58回日本白内障学会総会, 2019
【Original Research】
- Konno Y, Nagaoka S, Kimura I, Yamamoto K, Kagawa Y, Kumata R, Aso H, Ueda TM, Nakagawa S, Kobayashi T, Koyanagi Y, Sato K. New World feline APOBEC3 potently controls inter-genus lentiviral transmission. Retrovirology,15 (1), 31, 2018 (IF=3.7)
- Sato K, Misawa N, Takeuchi JS, Kobayashi T, Izumi T, Aso H, Nagaoka S, Yamamoto K, Kimura I, Konno Y, Nakano Y, Koyanagi Y. Experimental adaptive evolution of simian immunodeficiency virus SIVcpz to pandemic human immunodeficiency virus type 1 using a humanized mouse model. J. Virol., 92 (4), e01905-17, 2018 (IF=4.3)
- Krycer JR, Yugi K, Hirayama A, Fazakerley DJ, Quek L-E, Scalzo R, Ohno S, Hodson MP, Ikeda S, Shoji F, Suzuki K, Domanova W, Parker LB, Nelson ME, Humphrey SJ, Turner N, Hoehn KL, Cooney GJ, Soga T, Kuroda S, James DE. Dynamic Metabolomics Reveals that Insulin Primes the Adipocyte for Glucose Metabolism. Cell Reports, 21(12), 3536–3547, 2017 (IF=9.4)
- *Sano T, *Kawata K, *Ohno S, Yugi K, Kakuda H, Kubota H, Uda S, Fujii M, Kunida K, Hoshino D, Hatano A, Ito Y, Sato M, Suzuki Y, Kuroda S. Selective control of up-regulated and down-regulated genes by temporal patterns and doses of insulin. Science Signaling, 9(455), ra112, 2016 (IF=8.2) (*:co-1st authors)
- Okita Y, Shiono T, Yahagi A, Hamada S, Umemura M, Matsuzaki G. Interleukin-22-induced Antimicrobial Phospholipase A2 Group IIA Mediates Protective Innate immunity of Nonhematopoietic Cells against Listeria monocytogenes. Infect. Immun. 84(2), 573-579, 2015 (IF=3.4)
- Tokuyama K, Ohno S, Yoshikawa K, Hirasawa T, Tanaka S, Furusawa C, Shimizu H. Increased 3-hydroxypropionic acid production from glycerol by modification of central metabolism in Escherichia coli. Microb. Cell Fact., 13, 64, 2014 (IF=5.1)
- Nakashima N, Ohno S, Yoshikawa K, Shimizu H, Tamura T. A vector library for silencing central carbon metabolism genes with antisense RNAs in Escherichia coli. Appl. Environ. Microbiol., 80(2), 564–573, 2014 (IF=4.8)
- Ohno S, Shimizu H, Furusawa C. FastPros: Screening of reaction knockout strategies for metabolic engineering. Bioinformatics, 30(7), 981–987, 2014 (IF=6.9)
- Ohno S, Furusawa C, Shimizu H. In silico screening of triple reaction knockout Escherichia coli strains for overproduction of useful metabolites. J. Biosci. Bioeng., 115(2), 221–228, 2013 (IF=2.9)
- Matsuo M, Maeda T, Ono N, Sugihara S, Kobayashi I, Koga D, Hamasaki Y. Efficacy of Dextromethorphan and Cyclosporine A for Acute Encephalopathy. Pediatr. Neurol. 48(3), 200-205, 2013 (IF=4.2)
- 【Citation top10% paper】Saiki Y, Yoshino Y, Fujimura H, Manabe T, Kudo Y, Shimada M, Mano N, Nakano T, Lee Y, Shimizu S, Oba S, Fujiwara S, Shimizu H, Chen N, Nezhad ZK, Jin G, Fukushige S, Sunamura M, Ishida M, Motoi F, Egawa S, Unno M, Horii A. DCK is frequently inactivated in acquired gemcitabine-resistant human cancer cells. Biochem. Biophys. Res. Commun., 421(1), 98-104, 2012 (IF=3.6)
- Shimizu H, Horii A, Sunamura M, Motoi F, Egawa S, Unno M, Fukushige S. Identification of epigenetically silenced genes in human pancreatic cancer by a novel method “microarray coupled with methyl-CpG targeted transcriptional activation” (MeTA-array). Biochem. Biophys. Res. Commun., 411(1), 162-7, 2011 (IF=3.6)
【Review】
- Nakano Y, Aso H, Soper A, Yamada E, Moriwaki M, Juarez-Fernandez G, Koyanagi Y, Sato K. A conflict of interest: the evolutionary arms race between mammalian APOBEC3 and lentiviral Vif. Retrovirology, 14 (1), 31, 2017 (IF=3.4)
- Soper A, Juarez-Fernandez G, Aso H, Moriwaki M, Yamada E, Nakano Y, Koyanagi Y, Sato K. Various plus unique: Viral protein U as a plurifunctional protein for HIV-1 replication. Exp. Biol. Med. (Maywood), 242 (8), 850-858, 2017 (IF=2.4)
【Invited talks (including Japanese)】
- 大野 聡. 代謝システムの理解と応用に向けた数理解析. 日本生物工学会 バイオインフォマティクス相談部会 第一回勉強会, 2017
【Patent (including Japanese)】
- 中山 敬一, 清水 秀幸. 特願2018-157870 被検体に生じるイベントを予測するための判別器の生成方法、及び前記判別器を用いた被検体の層別化方法
【Original Research Written in Japanese 】
- 沖田 大和, 笹川稚依子, 関端広大, 吉田真奈美, 八田明, 井上秀雄. マウス IgE 介在性三相性皮膚反応の耳介腫脹に対する Glycyrrhetinic acid の効果. 薬理と治療, 42, 845-850, 2014
【Press release (including Japanese)】
【International conference】
- Ohno S, Quek LE, Krycer JR, Yugi K, Hirayama A, Soga T, James DE, Kuroda S. 13C-based dynamic metabolic flux analysis in insulin-stimulated adipocyte cell lines and systematic evaluation of regulation on the metabolism. Metabolic Engineering 12, Germany, 2018
- Nakano Y, Yamamoto K, Kumata R, Aso H, Soper A, Misawa N, Konno Y, Kimura I, Nagaoka S, Ikeda T, Juarez-Fernandez G, Ito J, Harris RS, Sato K, Koyanagi Y. Evolutionary journey of primate lentivirus transmission from chimpanzee to gorilla focusing on relationship between Vif and APOBEC3. Cold Spring Harbor meeting Retroviruses. New York, USA. 2018
- Ohno S, Krycer J, Yugi K, Quek LE, Hirayama A, Soga T, Kuroda S, James DE. Metabolic flux analysis in insulin-stimulated adipocytes form temporal patterns of intracellular metabolome data. Metabolic Engineering XI, Japan, 2016
- Tokuyama K, Ohno S, Yoshikawa K, Hirasawa T, Tanaka S, Furusawa C, Shimizu H. Increased 3-hydroxypropionic acid production from glycerol based on the genome-scale metabolic simulation in Escherichia coli. Metabolic Engineering X, Westin Bayshore, Canada, 2014
- Matsuzaki G, Okita Y, Umemura M. Innate protective immunity of non-hematopoietic cells against Listeria monocytogenes infection induced by IL-17 and IL-22. The 12 th Japan-Korea International Symposium on Microbiology (XII-JKISM), Japan, 2014
- Matsuzaki G, Okita Y, Shiono T, Hamada S, Umemura M. Mechanism of interleukin (IL)-17A-mediated enhancement of protective immunity against Listeria monocytogenes infection. 15th International Congress of Immunology, Italy, 2013
- Ohno S, Furusawa C, Shimizu H. FastPros: Efficient screening algorithm of gene knockouts for useful compound production using genome-scale metabolic model. KAIST-OSAKA U Symposium 2012, Korea Advanced Institute of Science and Technology, Korea, 2012
- Ohno S, Furusawa C, Shimizu H. FastPros: Screening method of multiple gene knockout for microbial production using genome-scale metabolic model. Metabolic Engineering IX, Le Bellevue Congress and Exhibition centre, France, 2012 【 Student/Young Investigator Poster Award / 若手研究者ポスター賞 受賞】
- Okita Y, Lee J, Nakamura K, Kobayashi M, Herndon D, Suzuki F. Improved production of human beta-defensins (HBDs) in grafted site skin tissues of human IMC/mouse chimeras treated with glycyrrhizin (GL). 9th The Annual Meeting of The American Association of Immunologists, Boston, USA, 2012
- Shimizu H, Fukushige S, Horii A. Identification of epigenetically silenced genes by the novel “methyl-CpG targeted transcriptional activation” method in human pancreatic cancer. AACR-JCA 8th Joint Conference, 2010
- Fukushige S, Shimizu H, Sato Y, Horii A. Identification of novel targets for aberrant methylation in pancreatic cancer by a newly developed method “methyl-CpG targeted transcriptional activation (MeTA)”. AACR 101st Annual Meeting 2010
【Domestic conference (including Japanese)】
- 清水 秀幸、中山 敬一. 人工知能を使って乳がん患者の生存予後を正確に予測する. 第41回日本分子生物学会年会, 2018
- 清水 秀幸、中山 敬一. 人工知能を使って乳がん患者の生存予後を正確に予測する. 第7回生命医薬情報学連合大会, 2018
- 大野 聡. インスリン刺激下の脂肪細胞における代謝フラックスとその調節. 新学術代謝統合オミクス 若手合宿, 2018
- Shimizu H, Matsumoto M, Nakayama KI. Artificial Intelligence Precisely Predicts Overall Survival of Cancer Patients. 第21回九州大学生体防御医学研究所リトリート, 2018 【 Best oral award / 最優秀口演賞 受賞】
- 中新井田 悠太, 半田知也, 川守田拓志, 岩田遥, 庄司信行. 片眼遮閉に伴う中距離及び近方視時の屈折値変化. 第 54 回日本眼光学学会総会, 2018
- Aso H, Ito J, Koyanagi Y, Sato K. Systematic investigation of transcriptional regulations of interferon-stimulated genes. 第66回日本ウイルス学会学術集会. 2018
- Konno Y, Nagaoka S, Kimura I, Yamamoto K, Kagawa Y, Kumata R, Aso H, Ueda MT, Nakagawa S, Kobayashi T, Koyanagi Y, Sato K. New World feline APOBEC3 potently controls inter-genus lentiviral transmission. 第66回日本ウイルス学会学術集会. 2018
- Sato K, Konno Y, Nagaoka S, Kimura I, Aso H, Kumata R, Ito J, Koyanagi Y. システムウイルス学的アプローチからを紐解くウイルスと宿主の進化的軍拡競争のダイナミズム. 日本進化学会第20回大会. 2018
- Aso H, Ito J, Suzuki Y, Sato K, Koyanagi Y. HIV-1感染ヒト化マウスにおける1細胞網羅的遺伝子発現解析. 第15回ウイルス学キャンプ in 湯河原. 2018
- Nagaoka S, Kawakami E, Ito J, Misawa N, Aso H, Nakaoka S, Dou Y, Satou Y, Shiroguchi K, Koyanagi Y, Sato K. 生体内のHIV-1複製におけるエピジェネティックネットワークの解明. 第15回ウイルス学キャンプ in 湯河原. 2018
大野 聡. 脂肪培養細胞の非定常フラックス解析および代謝制御の評価, 野生型および肥満モデルマウスにおける肝臓の代謝調節解析. 代謝モデリング勉強会, 2017
- 清水 秀幸、武石 昭一郎、中津海 洋一、中山 敬一. 固形がんにおけるFbxw7を標的とした静止期追い出し療法. 第40回日本分子生物学会年会, 2017
- 大野 聡, James R. Krycer, 柚木克之, Lake-Ee Quek, 平山明由, 曽我朋義, 黒田真也, David E. James. Systematic analysis of regulation on metabolism in insulin-stimulated adipocyte cell lines. CREST生命動態 領域会議, 2017
- Kimura I, Yamamoto K, Nagaoka S, Aso H, Konno Y, Nakano Y, Sato K & Koyanagi Y. Investigation of virus-host evolutionary arms race: APOBEC3 versus Vif. 第40回日本分子生物学会年会. 2017
- Nakano Y, Misawa N, Konno Y, Kimura I, Nagaoka S, Yamamoto K, Aso H, Kumata R, Juarez-Fernandez G, Soper A, Sato K, Koyanagi Y. Tracing the evolution of primate lentiviruses. 第40回日本分子生物学会年会. 2017
- Aso H, Yamamoto K, Kimura I, Konno Y, Nagaoka S, Nakano Y, Sato K, Koyanagi Y. Evolutionary arms race between APOBEC3 and lentiviruses in felids. 第65回日本ウイルス学会学術集会. 2017
- Nakano Y, Misawa N, Kimura I, Konno Y, Nagaoka S, Yamamoto K, Aso H, Sato K, Koyanagi Y. Influence of human APOBEC3H on primate lentivirus evolution. 第65回日本ウイルス学会学術集会. 2017
- Nagaoka S, Yamamoto K, Aso H, Kimura I, Konno Y, Nakano Y, Sato K, Koyanagi Y. ネコ科動物由来APOBEC3とネコ免疫不全ウイルスの進化的軍拡競争の解析. 日本進化学会第19回大会. 2017
- Nakano Y, Misawa N, Kimura I, Konno Y, Nagaoka S, Yamamoto K, Aso H, Juarez-Fernandez G, Soper A, Sato K & Koyanagi Y. ヒトAPOBEC3Hとレンチウイルスの攻防. 第20回日本レトロウイルス研究会夏期セミナー. 2017
- Soper A, Moriwaki M, Misawa N, Yamada E, Nakano Y, Aso H, Yoshikawa R, Sato K, Koyanagi Y. Evaluation of artificial HIV-1 heterogeneity in vitro and in vivo. 第20回日本レトロウイルス研究会夏期セミナー. 2017
- 大野 聡, James R. Krycer, 柚木克之, Lake-Ee Quek, 平山明由, 曽我朋義, 黒田真也, David E. James. Metabolic flux analysis in insulin-stimulated adipocytes from temporal patterns of intracellular metabolome data. CREST生命動態 第6回数理デザイン道場, 2016
- 清水 秀幸、武石 昭一郎、中山 敬一. ユビキチンリガーゼFbxw7を標的とした新規乳がん治療法の開発. 第39回日本分子生物学会年会, 2016
- 大野 聡, インスリン刺激下における脂肪培養細胞の非定常フラックス解析. 代謝モデリング勉強会, 2016
- Soper A, Misawa N, Yamada E, Nakano Y, Moriwaki M, Aso H, Yoshikawa R, Sato K, Koyanagi Y. Evaluation of artificial HIV-1 heterogeneity in vitro and in vivo. 第64回日本ウイルス学会学術集会. 2016
- 沖田 大和, 井石契子, 松原勤, 池田一雄, 河田則文,吉里勝利. 肝星細胞正常性維持へのサイトグロビン(CYGB)の関与. 第22回肝細胞研究会, 2015
- 沖田 大和, 松原勤, 池田一雄, 河田則文,吉里勝利. 肝星細胞によるサイトグロビンの発現に対する酸素分圧と基底膜成分の影響. 第41回肝臓学会西部会, 2015
- 大野 聡, 古澤力, 清水浩. 代謝シミュレーションを用いた、微生物によるものづくりのための多重遺伝子破壊探索手法の開発. 生命情報科学若手の会第5回研究会, 2014
- 徳山健斗, 大野 聡, 吉川勝徳, 古澤力, 清水浩. 代謝予測に基づいた3HP高生産大腸菌の構築. 日本農芸化学会2014年度大会, 2014
- 大野 聡, 古澤力, 清水浩. 微生物育種に向けた多重遺伝子破壊のin silicoスクリーニング. 生物工学若手研究者の集い(若手会)夏のセミナー2014, 2014
- 大野 聡, 古澤力, 清水浩. in silicoスクリーニングにより代謝物質生産が予測された大腸菌多重遺伝子破壊株の特徴解析. 生物工学会第65回大会, 2013
- 徳山健斗, 田中章太郎, 大野 聡, 吉川勝徳, 古澤力, 清水浩. 大腸菌によるグリセロールからの3-ヒドロキシプロピオン酸生産. 日本農芸化学会2013年度大会, 2013
- 清水 秀幸, C型慢性肝炎におけるテラプレビル3剤併用療法~当院における19例のまとめ~, 第196回日本消化器病学会東北支部例会, 2013
- 大野 聡, 微生物を利用したものづくりと細胞シミュレーションの応用. 八大学博士学生交流フォーラム第5回研究会, 2013
- 大野 聡, 古澤力, 清水浩, FastPros: 有用物質生産性向上に向けた多重遺伝子破壊のin silico スクリーニング. 生物工学会第64回大会, 2012
- 大野 聡, 古澤力, 清水浩. 代謝シミュレーションを用いた有用物質生産性を向上させる複数遺伝子破壊の予測アルゴリズム開発. 生物工学会第63回大会, 2011
- 大野 聡, 古澤力, 田中章太郎, 平沢敬, 清水浩. 大腸菌のゲノムスケール代謝モデルによる有用物質生産位向けた代謝予測. 生物工学会第62回大会, 2010
- 清水 秀幸, 福重 真一, 堀井 明. メチル化で発現抑制された遺伝子を特定する新規の方法「MeTA」の膵癌解析への応用, 第99回日本病理学会総会, 2010
【Guest column (including Japanese)】
- 清水 秀幸, 癌細胞はエクソソームを介してより転移能の高い細胞を模倣する. 細胞工学2015年9月号
- 清水 秀幸, 血糖降下・抗炎症作用を持つ内在性の新規脂質群の発見. 細胞工学2015年1月号
Contact Information
Any contacts should be directed to Prof. Hideyuki SHIMIZU.
- h_shimizu.dsc@tmd.ac.jp
- Department of AI Systems Medicine, M&D Data Science Center, Institute of Integrated Research, Institute of Science Tokyo
- Yushima 1-5-45, Bunkyo-ku, Tokyo 113-8510, JAPAN
